Featured Publications
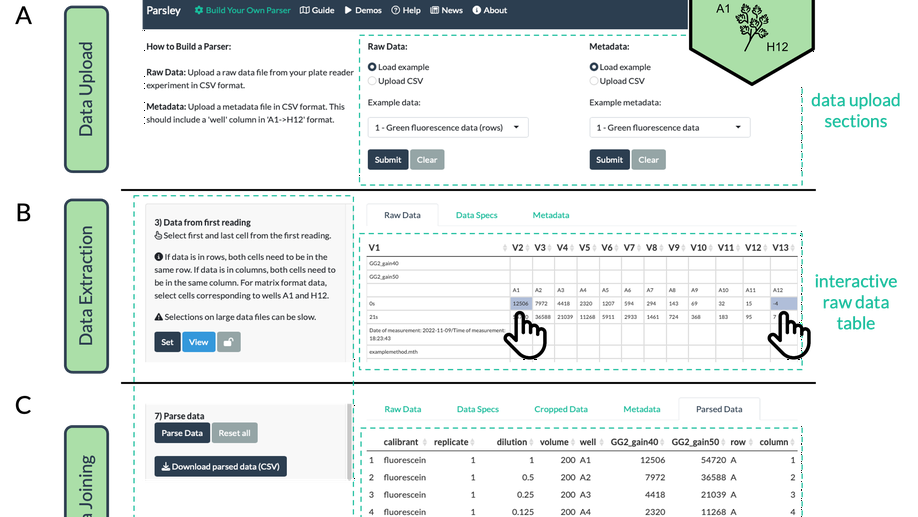
Parsley- a web app for parsing data from plate readers
As demand for the automation of biological assays has increased over recent years, the range of measurement types implemented by multiwell plate readers has broadened and the list of published software packages that caters to their analysis has grown. However, most plate readers export data in esoteric formats with little or no metadata, while most analytical software packages are built to work with tidy data accompanied by associated metadata. ‘Parser’ functions are therefore required to prepare raw data for analysis. Such functions are instrument- and data type- specific, and to date, no generic tool exists that can parse data from multiple data types or multiple plate readers, despite the potential for such a tool to speed up access to analysed data and remove an important barrier for less confident coders. We have developed the interactive web application, Parsley, to bridge this gap. Unlike conventional programmatic parser functions, Parsley makes few assumptions about exported data, instead employing user inputs to identify and extract data from data files. In doing so, it is designed to enable any user to parse plate reader data and can handle a wide variety of instruments (10+) and data types (53+). Parsley is freely available via a web interface, enabling access to its unique plate reader data parsing functionality, without the need to install software or write code.
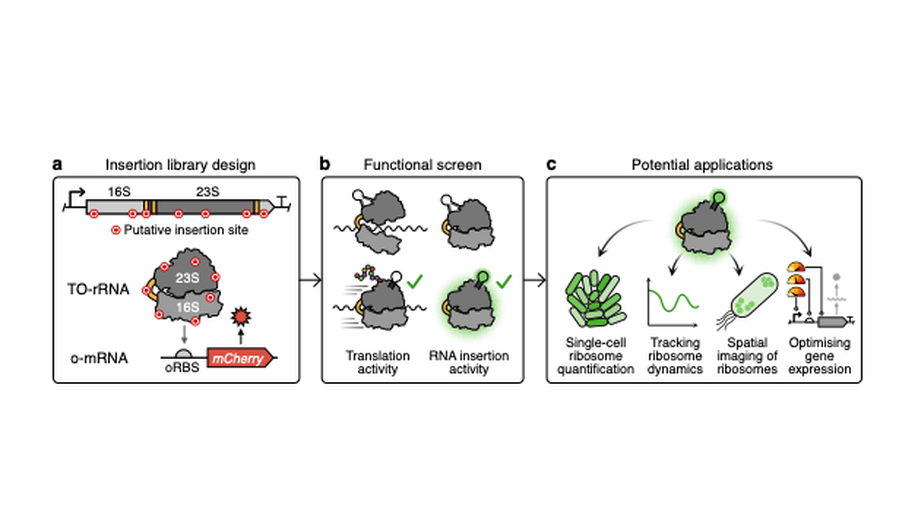
Engineering orthogonal ribosomes for real-time monitoring using fluorescence
A promising route to tackle the trade-off in cellular resources between synthetic protein production and cellular growth is to use a separate dedicated pool of orthogonal ribosomes to produce synthetic proteins. However, the optimisation of strains containing two ribosomal pools – native for the host cell’s proteome and orthogonal for synthetic proteins – has yet to be thoroughly explored. Here, we address this by creating orthogonal ribosomes that fluoresce by inserting fluorescent RNA aptamers into tethered orthogonal ribosomal RNA (TO-rRNA). To study the tolerance of the engineered ribosomes to aptamer insertion, we assembled and screened a library of candidate insertion sites, identifying several sites in both the 16S and 23S TO-rRNA that enables ribosome labelling with minimal effect on translation activity. Serendipitously, we identify one site in 23S TO-rRNA, where insertion appears to not only be tolerated but to enhance orthogonal ribosome activity, across multiple bacterial strains and RNA insertions. Using bulk and single cell assays, we demonstrate that this variant allows us to label orthogonal ribosomes for dynamic tracking and across populations, making it a promising tool for optimising orthogonal translation in engineered cells. Ribosome engineering offers great potential, both for the development of next-generation microbial cell factories, as well as a tool to expand our understanding of ribosome function in living cells. This work provides a window into the assembly, localisation and function of these molecular machines to meet these aims.
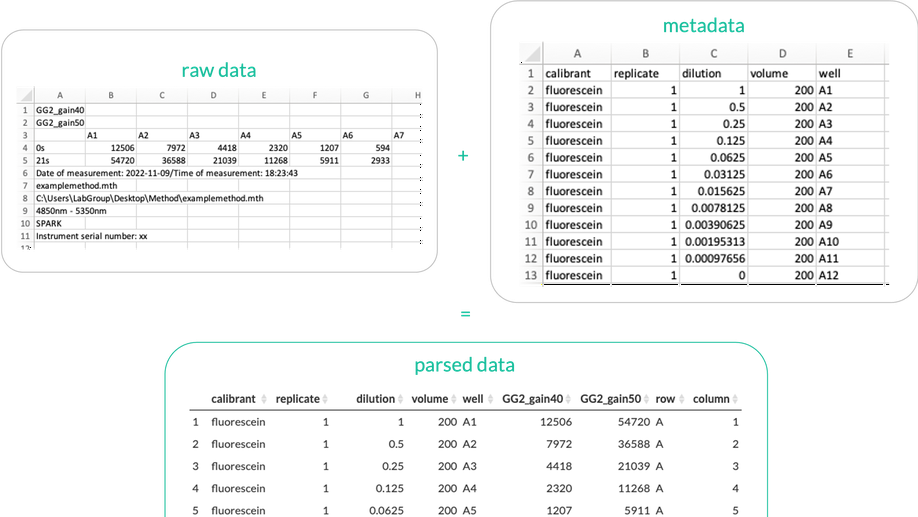
Parsley is a universal plate reader data parsing application.
Multiwell plate readers are an important tool in the life sciences. They are frequently used to measure fluorescence, absorbance and luminescence, among other measurement types. As they are heavily used for high-throughput assays and screens, their analysis benefits from automated (programmatic) data analysis. However, most plate readers export raw data in formats that software packages cannot work with, and that do not contain the necessary metadata for downstream analysis. A necessary initial step in every analysis is therefore extracting the data, reformatting it into the correct ‘tidy’ data structure, and joining it with any required metadata- we call this process ‘parsing’. While a few parser functions for certain export formats from certain plate readers have been written, there is no generic tool that can handle data parsing from any plate reader- this is why Parsley was built. More information is available on the GitHub page (https://github.com/ec363/parsleyapp) and app ( https://gbstan.shinyapps.io/parsleyapp ).
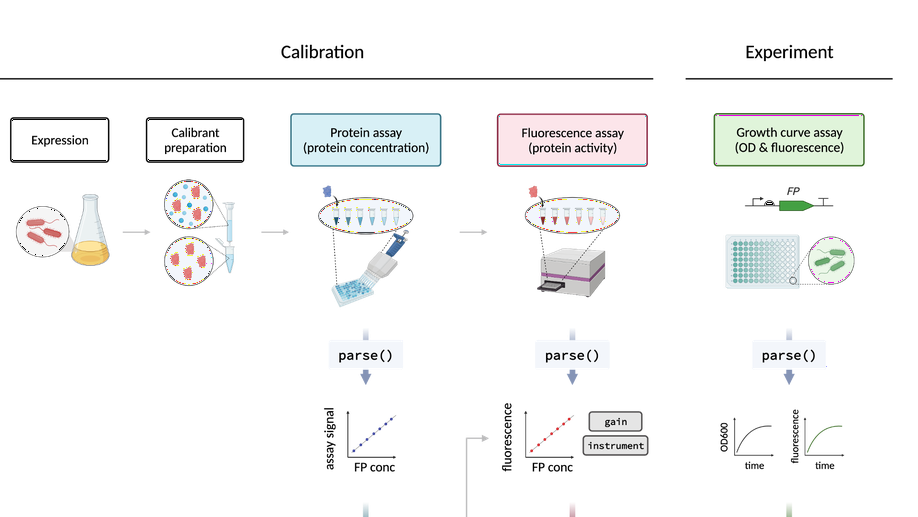
Absolute protein quantification using fluorescence measurements with FPCountR
This paper presents a generalisable method for the calibration of fluorescence readings on microplate readers, in order to convert arbitrary fluorescence units into absolute units. FPCountR relies on the generation of bespoke fluorescent protein (FP) calibrants, assays to determine protein concentration and activity, and a corresponding analytical workflow. We systematically characterise the assay protocols for accuracy, sensitivity and simplicity, and describe an ‘ECmax’ assay that outperforms the others and even enables accurate calibration without requiring the purification of FPs. To obtain cellular protein concentrations, we consider methods for the conversion of optical density to either cell counts or alternatively to cell volumes, as well as examining how cells can interfere with protein counting via fluorescence quenching, which we quantify and correct for the first time. Calibration across different instruments, disparate filter sets and mismatched gains is demonstrated to yield equivalent results. It also reveals that mCherry absorption at 600 nm does not confound cell density measurements unless expressed to over 100,000 proteins per cell. FPCountR is presented as pair of open access tools (protocol and R package) to enable the community to use this method, and ultimately to facilitate the quantitative characterisation of synthetic microbial circuits.
FPCountR- Fluorescent protein calibration for plate readers
The R package FPCountR enables the calibration of microplate readers using fluorescent protein calibrants. This enables users to monitor cellular fluorescent protein expression in absolute units. More information is available on the GitHub page (https://github.com/ec363/fpcountr) and website (https://ec363.github.io/fpcountr/ ).


